A brief overview of the current projects from our lab
I invite you to join us!
The Glickman Lab
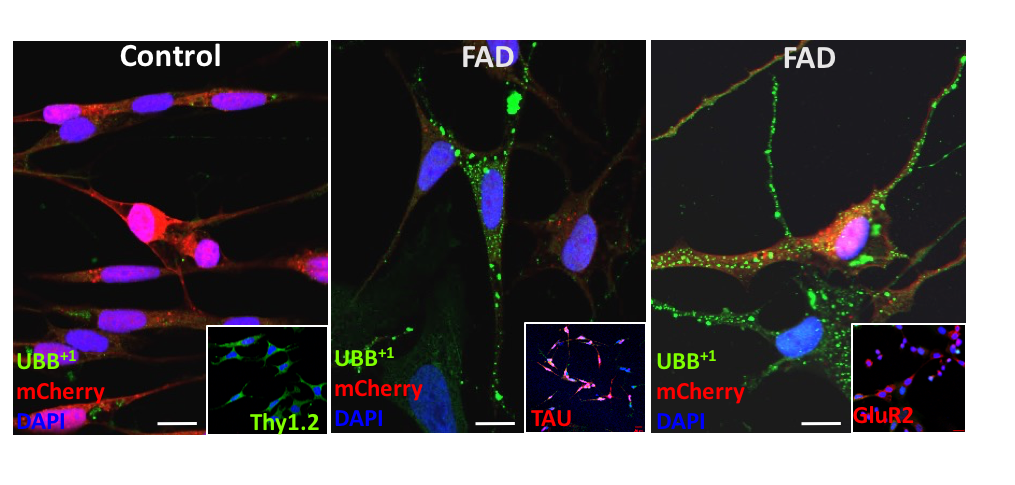
Involvement of protein turnover in the progression towards Alzheimer’s Disease in Neuronal models and attenuated protein turnover in ageing, Clearance of oxidized proteins and response to damaged mitochondria. The aim of our laboratory at the Technion is to understand the causative link between ubiquitin and Alzheimer’s disease. We want to understand if inefficient protein clearance is a risk factor for the development of sporadic Alzheimer’s disease, and if so, how it interferes with the healthy clearing mechanism of toxic protein clumps. What makes it appear sporadically is an enigma, which we aim to tackle by recreating the proper environmental and tissue context in the laboratory. We also plan to devise ways to block the formation of this mutant in stem cell-derived neurons in order to understand if this can stave off Alzheimer’s disease.
Proteasome structure and function, Mechanistic aspects of protein degradation by the ubiquitin-proteasome system. Our primary focus has been the dissection of the 19S regulatory particle of the proteasome into elementary functional units. Mapping subunit composition of the Lid and Base subcomplexes. Identification of a central unit within the Base that links substrate recruitment with proteolytic activation and the mechanistic role of the solenoid fold of the PC repeat (HEAT-like) regions of Rpn1 and Rpn2 that make up the central unit of the Base. How do the ATPases and the central Rpn1-Rpn2 unit in the Base control channel gating and substrate traffic into the 20S chamber? We are interested in dissecting the roles of the proteasome ATPases in the different steps of proteasome function.
Charting the cellular ubiquitin-linkage profile using Mass spectrometry and proteomics. The importance of proteolysis in homeostasis and regulation of the proteome and the specific recognition of ubiquitin and ubiquitin-like proteins. Under normal conditions, absolute quantification of conjugated ubiquitin by mass spectrometry demonstrates that a large portion of high molecular weight conjugated ubiquitin in whole cell extract is short chains or single ubiquitin modifications. Lysine48- and lysine63- linked chains of polyubiquitin are the most abundant linkage types naturally found in intact cells.
How do ubiquitination and proteasome-dependent-degradation participate in mitochondria function and dynamics? Biology of mitochondria membrane fusion and fission. In an exciting ongoing project, we are exposing a link between the ubiquitin-proteasome system and the mitochondria. By Ubiquitination and degradation of mitochondrial proteins, the ubiquitin-proteasome systems can influence mitochondria morphology and energy production in the mitochondria and viability life span. Components of the proteasome associated with the mitochondria. Mutants in Rpn11, a proteasome subunit resident in the Lid subcomplex display unique mitochondrial phenotypes. These mutants are also defective in proteolytic turnover of Fzo1, a dynamin-related trans-membrane GTPase embedded within the outer membrane of the mitochondria. We helped uncover direct evidence for the ubiquitination of Fzo1 at the mitochondria by the SCFMdm30 E3 ubiquitin ligase.