

Marie Curie Career Development Fellow (MSCA-IF)
Marie SkŁodowska-Curie Actions, Individual Fellowships, H2020-MSCA-IF-2016, European Commision
Indrajit Sahu completed his BSc in Zoology from Ravenshaw College, Cuttack, Odisha, India in 2006 and his MSc. in Microbiology from OUAT, Bhubaneshwar, Odisha, India in 2008. He then moved to ACTREC-Tata Memorial Centre, Navi Mumbai, India for his doctoral degree in 2010 and completed his Ph.D. in Life Science in 2015. For his first postdoc, he moved to Technion, Israel at Glickman’s Lab in 2015 and completed the project in 2017. He has also worked as an EC-Senior Researcher under MSCA-Individual Fellowship. Currently, he is an assistant professor at SRM- Institute of Science and Technology, Kattankulathur, Chennai.
Email: indrajis@srmist.edu.in
https://www.researchgate.net/profile/Indrajit_Sahu
Project Title: Unraveling the mechanism of Ubiquitin-dependent and independent protein degradation by Proteasomes: A mechanistic approach.
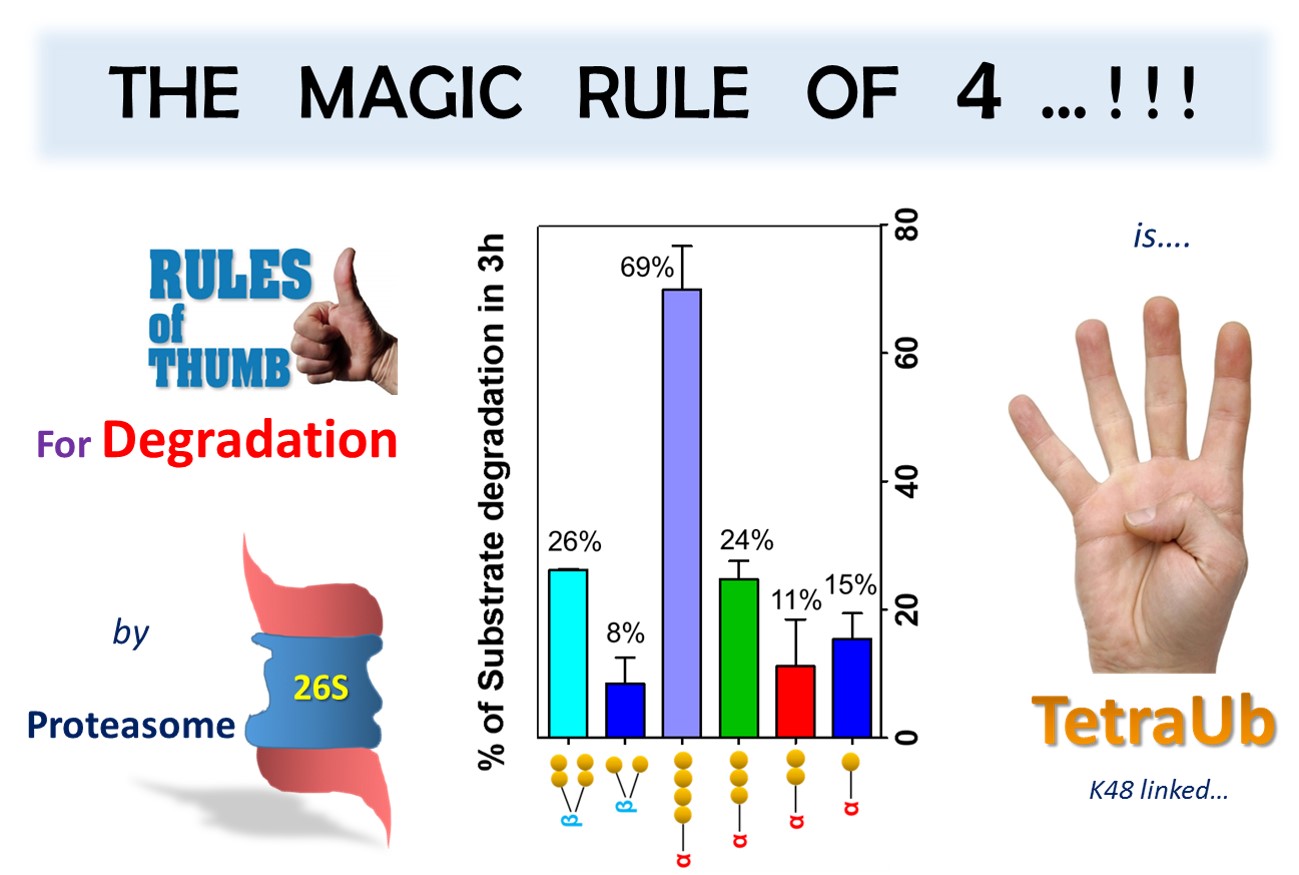
The Ubiquitin Proteasome System (USP) is the central machinery in eukaryotic cells for clearing the mess of old, unused or damaged proteins in a highly regulatory fashion. Many reports addressed; how ubiquitin chain length, linkage type and its susceptibility to deubiquitinases affect substrates processing by 26S proteasome. However, the precise ubiquitination requirements for substrate degradation by proteasome remain ambiguous. In order to solve this riddle, we carefully synthesized a set of K48-linked polyUb-globin conjugates differing in ubiquitin chain length, position and susceptibility to deubiquitinases. By studying the fate of each substrate incubated with 26S proteasomes isolated from human tissue, and monitoring products generated we concluded (1) the cleavage of the proximal isopeptide bond is not a prerequisite for proteasomal degradation; (2) by overcoming trimming at the proteasome, tetraUb is a fundamentally different signal than shorter chains, and (3) the tetra-ubiquitin chain can be degraded with the substrate.
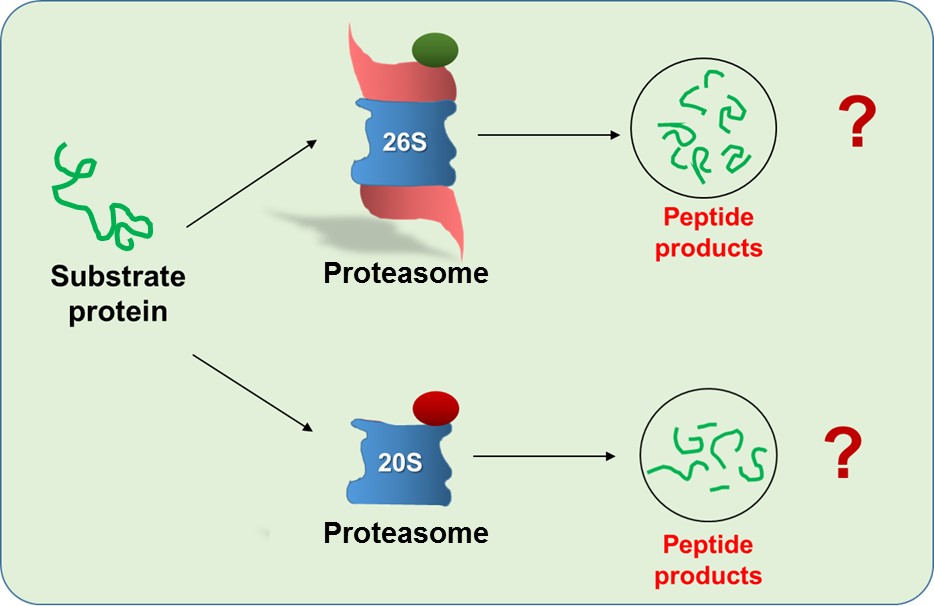
Proteins targeted for degradation are either ubiquitinated or non-ubiquitinated; however, they are processed in the 20S proteasome core, cleaved into small peptides and released. Some peptides escape further hydrolysis and are loaded onto MHC class I as a “self” signal to CD8-T-cells. Therefore, the cleavage specificity by proteasomes consequently determines efficiency of the immune response. However, it is still unclear how ubiquitination affects substrate processivity by different specifies of proteasomes which was the main focus of our current study. By choosing an unstructured substrate, we could compare proteolytic outcomes from different human proteasome species (20S and 26S) of highly homogenous synthetic non-ubiquitinated and ubiquitinated conjugates. Furthermore, we are trying to recapitulate this phenomenon in cellular model systems that mimic pathological conditions.
Publications:
Ding Z, Xu C, Sahu I, Wang Y, Fu Z, Huang M, Wong CCL, Glickman MH, Cong Y. Structural Snapshots of 26S Proteasome Reveal Tetraubiquitin-Induced Conformations. Mol Cell. 2019 Feb 12. pii: S1097-2765(19)30038-3. doi: 10.1016/j.molcel.2019.01.018. [Epub ahead of print] https://www.cell.com/molecular-cell/fulltext/S1097-2765(19)30038-3
Tiwari R, Sahu I, Soni, Sathe G.J, Thapa P, Patel P, Sinha S, Vadivel C, Patel S, Oak S, Thorat R, Gowda H, and Vaidya M.M. (2018). Depletion of Keratin 8/18 modulate oncogenic potential of A431 cells by governing multiple signaling pathways. FEBS J. Feb 2018, Vol. 285, Issue 4. DOI: 10.1111/febs.14401. https://febs.onlinelibrary.wiley.com/doi/full/10.1111/febs.14401
Tiwari R, Sahu I, Soni B, Sathe G.J, Datta K.K, Thapa P, Dhaka B, Sinha S, Gowda H, and Vaidya M.M. (2017). Quantitative phosphoproteomic analysis reveals system-wide signaling pathways regulated by site-specific phosphorylation on Keratin-8 in skin squamous cell carcinoma derived cell-line. Proteomics. 10.1002/pmic.201600254. http://onlinelibrary.wiley.com/doi/10.1002/pmic.201600254/full
Singh S.K, Sahu I, Mali S.M, Hemantha H.P, Kliefeld O, Glickman M.H, and Brik A. (2016). Synthetic uncleavable ubiquitinated proteins dissect proteasome deubiquitination and degradation, and highlight distinctive fate of tetraubiquitin. Am. Chem. Soc. December-6, 2016. 138, 16004−16015. DOI: 10.1021/jacs.6b09611. http://pubs.acs.org/doi/full/10.1021/jacs.6b09611
Hjerpe R, Bett J.S, Keuss M.J, Solovyova A, McWilliams T.G, Johnson C, Sahu I, Varghese J, Wood N, Wightman M, Osbourne G, Bates G.P, Glickman M.H, Trost M, Knebel A, Marchesi F, and Kurz T. (2016). UBQLN2 connects the proteasome with the HSP70 system to promote autophagy-independent protein aggregate clearance. Cell, Vol. 166, 1–15, August 11, 2016. http://www.cell.com/cell/fulltext/S0092-8674(16)30866-2
Sahu I, Sangith N, Ramteke M, Gadre R, Venkatrama P. (2014). A novel role for the proteasomal chaperone PSMD9 and hnRNPA1 in enhancing IκBα degradation and NF-κB activation – functional relevance of predicted PDZ domain–motif interaction. FEBS J. Vol. 281 (11), 2688-2709. http://onlinelibrary.wiley.com/doi/10.1111/febs.12814/full
Sangith N, Srinivasaraghavan K, Sahu I, Desai A, Medipally S, Somavarappu A.K, Verma C, Venkatraman P. (2014). Discovery of novel interacting partners of PSMD9, a proteasomal chaperone: Role of an Atypical and versatile PDZ-domain motif interaction and identification of putative functional modules. FEBS Openbio. Vol 4, 571–583. http://onlinelibrary.wiley.com/doi/10.1016/j.fob.2014.05.005/full
Academic Awards:
Funding:
https://ec.europa.eu/research/participants/portal/desktop/en/home.html
